A disk impregnated with erofloxacin antibiotic has inhibited the growth of a "lawn" of Gram-negative bacteria growing on an agar plate. The diameter of the area where growth is inhibited is a measure of either the antibiotic's effectiveness or bacteria's resistance to the antibiotic. In this case, the antibiotic is inhibiting growth of the bacteria isolated from a shark.
Evaluating Antimicrobial Resistance
in Wildlife and Fisheries
Mark A. Mitchell
Assistant Professor and
Director
Louisiana State University Wildlife Hospital
Co-Principal Investigator
Antimicrobials are substances capable of destroying or inhibiting the growth of disease-causing microorganisms. Some antimicrobials, such as antibiotics (e.g., penicillin or streptomycin), are derived from certain fungi, bacteria, and other organisms. Other antimicrobials are synthesized in pharmaceutical laboratories.
The advent of antimicrobials during the twentieth century was one of the most significant contributions to humanity. Historically, bacterial infections were responsible for a significant number of diseases and deaths in both developed and underdeveloped countries. Once antimicrobials came on the scene, however, people had an effective weapon at their disposal. The desire to improve on the types and availability of antimicrobials has led to the development of a multi-billion-dollar pharmaceutical industry.
Prescription for trouble?
There is a downside to this trend. The
availability of these compounds, together with a desire to promote and
sell products, has caused antimicrobial usage to skyrocket. Physicians
and veterinarians routinely prescribe antimicrobials to cover a broad
spectrum of pathogens without isolating a pathogen or confirming an antimicrobial-sensitivity pattern for a pathogen. This inappropriate use of antimicrobials has been identified as
a source of antimicrobial resistance in microbes found
in humans and animals.
The occurrence of antimicrobial-resistant bacteria in human and veterinary hospitals has generated interest among health professionals, because the development of “super-bacteria” presents a special health risk to people in these facilities. Nosocomial infections (infections with antimicrobial-resistant microbes in a hospital or hospital-like setting) are responsible for over 100,000 human deaths per year. They are also a concern in veterinary hospitals, with resistant microbes being found on common surfaces.
Additionally, veterinarians have used antimicrobials to promote growth and weight gain in certain species of livestock. This management technique may result in human exposure to antimicrobial-resistant microbes through the food chain.
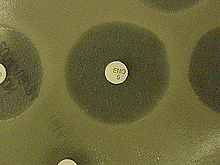
This image shows the results of a Kirby-Bauer disk diffusion antibiotic-susceptibility test. Antibiotic-impregnated disks (containing various antibiotics) are placed onto the agar surface, which contains a "lawn" of growing bacteria isolated from a bull shark caught in Louisiana waters. As the bacteria on the lawn grow, the antibiotic diffusing from the disks inhibits its growth varying degrees. As indicated by the varying zones of growth inhibition surrounding each disk, the Gram-negative bacteria isolated from the shark are resistant to some of the antibiotics in the disks at the concentrations tested. The diameter of the inhibition zone identifies the bacteria as "susceptible," "intermediate," or "resistant" to specific concentrations of an antibiotic. Click image for larger view.
Resistance in the wild
Only a few studies have evaluated wildlife and fisheries for antimicrobial-
or antibiotic-resistant bacteria. In one study, Salmonella bacteria
found in clinically diseased wild birds (i.e., owls and goldfinches) showed resistance
patterns to the synthetic antibacterial compound sulfamethoxazole. In another, investigators
identified widespread antimicrobial resistance in a population of starlings
in Iran. All 19 birds captured in that study had at least
one bacterium that had antimicrobial resistance. The antimicrobial
resistance patterns identified in the starlings were similar to those
described for chickens. Studies conducted at Louisiana State University
(LSU) found wild raptors (birds of prey) that harbored microbes resistant
to important synthetic antibiotics, such as ciprofloxacin and enrofloxacin.
Preliminary research at LSU suggests that antimicrobial resistance occurs in marine environments, as well. Bacteria with antimicrobial-resistance patterns to many different synthetic and natural antibiotics were found on swabs collected from the cloaca (the common intestinal and urogenital opening) of sharks in Belize, Massachusetts, and Louisiana.
How bacteria develop resistance
Understanding how bacteria develop resistance is important to understanding
how to control resistant bacteria. Antimicrobial resistance may
occur as a result of acquired resistance or evolution. Acquired resistance
frequently occurs as a result of the transfer of genetic material from one
bacterium to another by conjugation, transduction, transformation, and
plasmids, or by mutations from exposure to environmental mutagens. Mutation
in the resistant bacteria may occur at a rapid rate, because the logarithmic
growth rates of bacteria lead to rapid generation times. With the
advent of molecular technologies, it is now possible to determine if
bacteria are selecting for resistance naturally or through acquired resistance. Today, we can now look at the DNA of the bacteria to determine how the resistance
is occurring.
Environmental sentinels
The findings of these and ongoing studies at LSU
suggest that wildlife and fisheries may be important environmental sentinels
for antimicrobial resistance. Because many terrestrial and marine
species are migratory, they may also spread resistant microbes to previously
unaffected environments. On the Estuary to the Abyss Expedition,
we will attempt to sample fishes from relatively pristine environments to
determine if antimicrobial resistance is also occurring in environments
considered to be far removed from human influence.
References:
- Adetosoye AI. Transmissible drug resistance in human and animal strains of Enterobacteriaceae and Pseudomonas aerations. Res Vet Science 1980; 29: 343-345.
- Hudson CR, Quist C, Lee MD, Keyes K, Dodson SD, Sanchez S, White DG, Maurer JJ. Genetic relatedness of Salmonella isolates from nondomestic birds in Southeastern United States. J Clin Microbiol 2000; 38: 1860-1865.
- Mitchell MA, Roy A, Vennen K, Heatley JJ, Tully TN. Antibiotic resistance patterns of microbes isolated from raptors from Louisiana. Proc Assoc Avian Veterinarians 2001: 227-228.