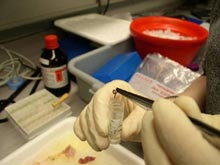
A researcher extracts DNA from this parasitic nematode that lives in association with Antarctic fish. Click image for larger view.
Genomics in Deep-sea Microbiology: What's Down There? What Are They Doing?
Naomi Ward
Assistant Investigator
The Institute for Genomic Research
What is genomics? Genomics is the study of an organism’s genome, or genetic material. The genome contains the coded instructions necessary for the organism to build and maintain itself. In most organisms, the genome consists of deoxyribonucleic acid, a big name for something we've commonly come to know as DNA.
Here's a little background on DNA. It is composed of four building blocks. (These are the nucleotides, which are also known as bases). The particular order of these building blocks -- in a string of DNA -- is a code for the production of proteins. Proteins are key structural components of the cell. They also act as enzymes, or chemical catalysts, for numerous processes that are essential to the cell.
Genomic Technologies and Bacteria
Genomic technologies, the methods we use to study genetic material, have many useful applications. For one, they can help us determine the identity and activities of bacteria living in diverse environments, including the deep-sea habitats we will visit on the 2004 Gulf of Alaska Seamount Expedition.
Bacteria are key players in the marine environment. In a drop of seawater, there can be anywhere from a million to a billion bacterial cells. Similarly, high numbers of bacteria are found in marine sediments, even far beneath the ocean floor. These microbes have important roles in the cycling of essential elements, such as carbon, nitrogen, phosphorus, and sulfur. They serve as a food source for other forms of marine life, and participate in the decay process that regenerates nutrients from dead organisms. Recent genomic studies have shed light on the structure and function of these marine bacterial communities and have even revealed the occurrence of a new type of bacterial photosynthesis in the oceans.
Sampling the Seamounts
During this expedition, we will sample marine bacteria from deep water, surface water, deep sediments, and biofilms (layers of bacterial cells) from the exterior surfaces of animals. We will use the deep submergence vehicle (DSV) Alvin to collect a five-bottle set of water samples. The Alvin sub pilot will trigger the water collection at different sites. An alternative device for deep-water collection is the CTD (Conductivity-Temperature-Depth) package, which carries 23 larger bottles that are triggered to collect water at different depths. Once on board, the samples will pass through a series of filters to trap microbes of different sizes.
Deep-sea sediment samples are captured by devices called “push corers.” Attached to Alvin, push corers are hollow tubes that are pushed into the mud. Back on the ship, the push cores (the tubes' contents) can be sliced up so that we have separate sediment samples from different depths.
We will also collect biofilm material from the outside of animals. A biofilm is a layer of bacterial cells held together on a solid surface that can be either living (e.g., a crab) or nonliving (e.g., a rock). The bacteria produces a mucus-like "glue" that holds the biofilm together. Biofilms are found everywhere, in both natural and human-made settings. They can pose a threat to human health, such as when a biofilm forms on an indwelling medical device (e.g., a catheter). In contrast, the biofilms that form on deep-sea surfaces may actually help other larger organisms, such as larvae, attach to and colonize that surface.
On our 2002 expedition to the Gulf of Alaska seamounts, we became particularly interested in the microbes living on the surface of deep-sea octocorals. We observed that different coral species had quite different associated microbial populations. This suggests that perhaps these populations are specialized to be beneficial to the coral, either by providing nutrients or by protecting the coral from other disease-causing microbes (Phylogenetic Diversity in Microbial Communities Associated with Gulf of Alaska Deep-sea Corals PDF, 58K).
When return to the Gulf of Alaska seamounts this August, we will take many more samples (including multiple samples of each individual species) than we did in 2002. This will allow us to be more confident about our conclusions regarding the microbes associated with each coral.
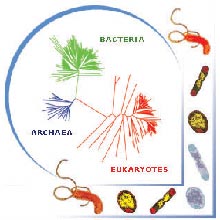
Genomics is helping us to understand better the evolutionary relationships of all living things: bacterial, archaeal, and eukaryotic. Click image for larger view.
Back at the Lab
During and after the cruise, scientists will extract and purify DNA from the samples. To determine which bacteria are present, we will sequence a particular gene (the 16S ribosomal RNA -- 16S rRNA -- gene) that all bacteria possess and that we can use as a “marker” gene. If there are 500 different bacteria in a given sample, we will get 500 different 16S rRNA gene sequences. We can identify these by comparing them to a database containing all known sequences from this gene.
To determine what these bacteria are doing in their habitat, we will sequence larger pieces of DNA, using the same methods that we apply to sequencing whole genomes of cultured bacteria. Comparison of these sequences to the genomic databases will allow us to assign a tentative function to each sequence.
Applications
The ultimate goal of applying genomic approaches to studying deep-sea bacterial communities is to identify which bacteria are present and what they are doing. If we can determine which proteins are being produced, we can reconstruct the pathways of metabolism for these different bacteria. We will know what substrates (food-stuffs) they use for nutrition, and how they bring them into their cell and break them down. We may also catch a glimpse of how bacteria fight each other and defend their territory, through information from genes that encode antibiotics and other substances.
The genomic information captured may also reveal the presence of bacteriophages (parasitic viruses). These viruses, which prey on bacteria, often insert their own DNA into the bacterium’s DNA. (This is part of the the viruses' sneaky strategy to take over the bacterium’s replication machinery and make thousands of copies of themselves.) Bacteriophages are thought to be a key force in shaping the structure of natural bacterial communities. Additional information on their distribution and association with particular bacteria may help us understand this process.