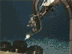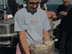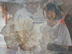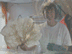Fig. 1. A large Lophelia coral bush with a squat lobster (Eumunida picta) and sea urchin (Echinus, below the crab).Click image for larger view and image credit.
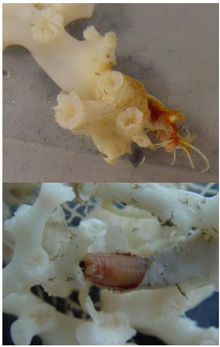
Fig. 2. Two views of a eunicid polychaete worm ‘at home’ on Lophelia pertusa coral. Click image for larger view and image credit.
Community Genetics - All About Relationships
Dr. Cheryl Morrison
Biologist
USGS-BRD, Leetown Science Center, Aquatic Ecology Branch
Deep Reef Fauna
When you picture a coral reef, you likely envision a complex coral landscape in clear warm water with a high biodiversity including colorful fishes and invertebrates. Although tropical coral reefs receive a lot of attention from both scientists and enthusiasts, our understanding of the physical, chemical, and biological forces that shape these ecosystems remains limited.
Now imagine coral reefs that occur over a thousand feet below the ocean’s surface (see World of Deep Sea Corals essay). Through research using submersibles and remote vehicles, deep-sea reefs dominated by the coral Lophelia pertusa have been documented along the Southeastern US and Gulf of Mexico continental slopes, as they have been in other areas of the world. Like their shallow-water, tropical counterparts, Lophelia reefs appear to have a taxonomically diverse cast of invertebrate characters that call such deep reefs ‘home’. A particularly visible invertebrate is the squat lobster, Eumunida picta, that can often be found perched on top of Lophelia bushes. Other inverts such as the sea urchin, Echinus, and a eunicid polychaete usually occur in this habitat as well. See essay by Nizinski on the invertebrates.
Repeated observations of invertebrates co-occurring on Lophelia reefs leads one to ask questions regarding the ecological significance of these associations, such as “How much do Lophelia-associated invertebrates depend upon the corals to survive?”, “Do the corals also rely on the invertebrates to remain healthy and build reefs?”, and ultimately, “Why are they found together?” On this mission, data regarding relative abundances, behaviors, and spatial distributions of Lophelia invertebrates will be gathered, yet answering broad ecological questions remains difficult without additional means to compare patterns across latitudes, depths and longer time frames.
Molecular Toolkit- Defining ‘Invisible’ Boundaries
A relatively new set of molecular techniques can be applied to coral and invertebrate tissue samples taken on deep-sea cruises in order begin to answer broad ecological questions like those posed above. Through a comparison of variability in DNA fragments from populations of each species geographically distributed along the Southeastern US, a clearer picture of genetic connectivity will result. We begin with a null hypothesis that Lophelia and the invertebrates occur together by chance. If this is true, we would expect to find different patterns of genetic variation for each species. If, however, the species are closely tied and relate to environmental and oceanographic features similarly, we may see shared patterns of genetic differentiation between Lophelia and the associated invertebrates among distant reefs. Or in other words, we may see ‘breaks’ or ‘gaps’ in the genetic continuity (gene flow) between distant reefs that are shared between corals and their associated invertebrates. This could mean that there is a co-evolved community whose members over thousands of years have responded to environmental pressures in similar ways. A regional genetic pattern shared across species is called ‘genealogical concordance’ and can be interpreted as shared historical elements in either evolutionary or ecological factors. Once patterns among taxa are deciphered, we may look to other types of data taken about the study sites to see whether there are correlations with physical, chemical or topographic conditions.
The molecular tools used to assess genetic continuity vary according to organism. Mitochondrial DNA sequences have typically been used to assess regional patterns in animals and will be used for our work on the squat lobster, urchin, and polychaete. Coral mitochondrial DNA evolves at a much slower rate, however, and often does not show differences between species. Therefore, we have developed more sensitive microsatellite markers for southeastern U.S. Lophelia. Microsatellite markers are short repeats of DNA nucleotides, such as ‘ATATAT’ or ‘GATAGATA’ that occur frequently in the nuclear genome. The number of copies of the repeat changes very quickly. An individual will receive one copy of this region of DNA from each of its parents, or in other words, will have 2 copies. We assess what copies are present in a piece of coral by separating out the DNA fragments by size. Different repeat numbers will result in different sized fragments. If both fragments are the same size, there will be only one ‘allele’ that is visible, and we assume both copies are the same (a homozygote). If there are two different sized alleles, the individual is a heterozygote. When many of these regions are examined in many animals, we obtain a detailed and accurate picture of the genetic structure. Such an analysis will allow for a fine-scale assessment of relationships within and between reefs along the southeastern US continental slope (Cape Lookout, NC to the central Gulf of Mexico). Tissues will be preserved while at sea, and the genetic analyses will take place in the USGS genetics lab in West Virginia.
In addition to these broad, multi-species relationship questions, our genetics data will allow us to examine the basic taxonomy of the corals and some of the invertebrates. We are exploring a frontier region that is poorly known. This means that what we currently know about taxonomy of these organisms is very likely subject to change. Many of these species have never been studied with modern genetic techniques. We will address such questions as: Are all of the Lophelia colonies in a given area clones? Do the corals we sample belong to accurately described species or do they belong to a different species (perhaps new to science)? What kind of population structure does a wide-spread coral species, like Lophelia, exhibit?

Fig. 3. Microsatellite alleles at the A5 locus for several Lophelia samples. The number of repeats of each ‘AAC’ allele is determined from the position of the peak, with larger fragments (more repeats) further to the right. Individuals shown are all heterozygotes since they have two peaks. Many loci are surveyed and a multilocus genotype is obtained for each sample. Click image for larger view and image credit.
Keeping Reefs Healthy
From this work, information regarding past and present connectivity among reefs for both the coral and some of its key reef inhabitants will be gained. Likewise, inferences can be drawn about larval dispersal for these organisms, which is presently unknown for most. Such data will be important to consider when attempts are made to protect or recover deep-sea Lophelia reefs. For example, if a reef is destroyed by fishing gear, oil exploration or other human activities, whether recovery is likely to come from local corals and invertebrates or from those transported a long distance is critical to the success of the recovery. Protected areas could be designed with these limits in mind. Also, more will be known about the physical factors that are important to reef development and existence, which should also be useful in effective management practices.