
by James Murphy, NOAA Office of Ocean Exploration and Research
Cheryl Morrison, Expedition Science Co-Lead, U.S. Geological Survey, Leetown Science Center
June 30, 2018

Sponges, corals, and other invertebrates live hundreds of meters below the sea surface. Image courtesy of the NOAA Office of Ocean Exploration and Research, Windows to the Deep 2018. Download larger version (jpg, 1.2 MB).
Although NOAA’s Office of Ocean Exploration and Research (OER) and partners have the ability to send a remotely operated vehicle (ROV) to the seafloor to explore and document what organisms may inhabit the deep sea, ROV Deep Discoverer (D2) is limited in the area in which it can survey. Outside of the ring of light D2 projects, there lies an unexplored world of organisms either just beyond the ROV’s reach or avoiding its whirring operating sounds and bright lights. Due to these shortfalls in exploration capabilities, we could be missing undiscovered cephalopods, new species of fish and sharks, and nearby sponges or corals that are just out of sight on each dive. However, advancements in molecular biology have provided a means for conducting metaphorical roll calls for deep-sea organisms during future exploration missions.

Remotely operated vehicle Deep Discoverer’s limited field of view on the seafloor below Seirios during Dive 04 of the Windows to the Deep 2018 expedition. Image courtesy of the NOAA Office of Ocean Exploration and Research, Windows to the Deep 2018. Download larger version (jpg, 294 KB).
All organisms leave behind a DNA footprint in their surrounding environment, whether they move through the area or simply grow in a stationary manner. Scales, mucus, feces, and other DNA sources contribute to what is commonly referred to as eDNA, or environmental DNA, which is DNA that can be collected from sampling environments such as seawater, freshwater, or sediment. Scientists employing eDNA have used it to track the spread of invasive species, monitor endangered animals, as well as learn more about the environment in hard to access areas. Improvements in DNA collection, preservation, processing, and analysis protocols have improved the reliability of identifying organisms through eDNA sampling and decreased the cost of DNA identification of different species by streamlining broadscale organism identifications.
The overall challenges of deep-sea exploration (remoteness, sophisticated technology requirement, and expense) mean that understanding of biological community and food web structure within various deep-sea environments and connectivity across ocean-basin scales is severely limited as compared to what we know about shallow water and terrestrial ecology. Thus, the integration of eDNA sampling is highly valuable, as scientists across the globe pine to learn more about the distribution of organisms within these unique ecosystems.

Once aboard NOAA Ship Okeanos Explorer, water collected in niskin bottles during the Windows to the Deep 2018 expedition Dive 04 is transferred to sterile containers for later eDNA processing. Image courtesy of the NOAA Office of Ocean Exploration and Research, Windows to the Deep 2018. Download larger version (jpg, 8.5 MB).
During Windows to the Deep 2018: Exploration of the Southeast U.S. Continental Margin expedition, we are currently conducting a technology demonstration of eDNA sampling, to assess the newly developed methodology during the exploration of areas off the shores of Florida, Georgia, and North and South Carolina. By collecting seawater during ROV dives, scientists can greatly enhance the sensing of cryptic animals in the deep-sea through the application of eDNA analysis for identification of presence/absence of species.
Sampling of eDNA allows for species detection and evaluation at a scale far greater than current strategies of sampling, as it allows for detection of organisms that the ROV may never have even seen. Additionally, sampling with D2 is often limited largely to sessile organisms. However, one eDNA water sample has the potential to contain the DNA of tens, if not hundreds, of organisms that would otherwise go unobserved, thereby amplifying the amount of information collected about the habitat being characterized.

Sponges, corals, urchins, and other organisms populate outcrops of hard substrate on the seafloor. Image courtesy of the NOAA Office of Ocean Exploration, Windows to the Deep 2018. Download larger version (jpg, 1.2 MB).
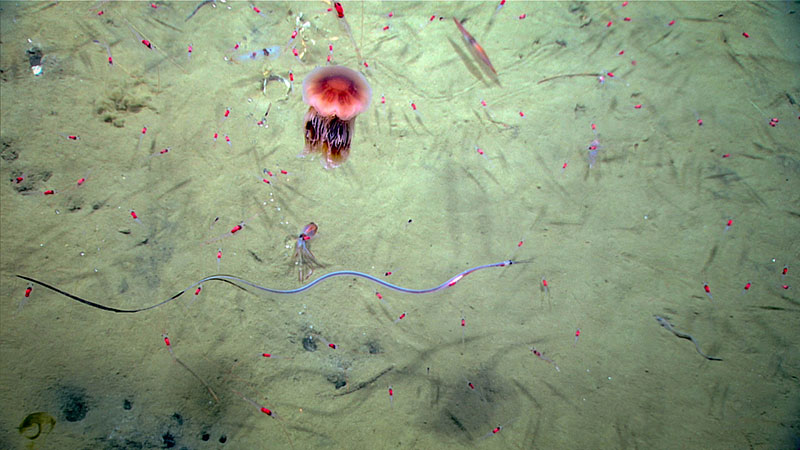
Many different organisms, including a snipe eel, jellyfish, shrimp, and squid, seen in the water during Dive 16 of the Windows to the Deep 2018 expedition. Image courtesy of the NOAA Office of Ocean Exploration, Windows to the Deep 2018. Download larger version (jpg, 578 KB).
Data resulting from this expedition are anticipated to inform future eDNA efforts, provide explorers and researchers with improved insight into deep-sea biological community structure in this region, and provide opportunities to improve upon sampling and processing methods for ROV-based eDNA collections. Such information could have incredible impacts, greatly improving our understanding of what lives in the deep sea.