
By Eric Collins and Sarah Hardy, Expedition Principal Investigators, University of Alaska Fairbanks
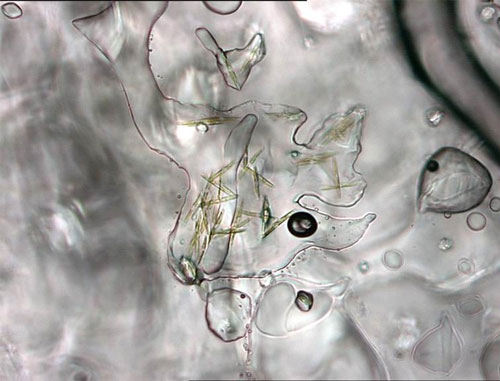
Single-celled (unicellular) algae, which develop in the lowermost sections of sea ice, often forming chains and filaments. Ice algae are an important component of the Arctic marine food web. Image courtesy of the NOAA Arctic Research Program. Download larger version (jpg, 123 KB).
The Arctic Ocean is home to an extraordinary diversity of life, including the unseen majority: microbes. Algae, heterotrophic protists, bacteria, archaea, and viruses all live together in a hidden ecosystem that recycles nutrients within Arctic seawater, sediment, and sea ice, making possible the amazing bounty of macroscopic life found there.
Though this microscopic ecosystem is usually hidden from our view, new technologies allow us to paint a much more accurate portrait of these microbes than has ever been possible before. As we explore these communities, we learn about how they survive in extreme environments, we discover their roles in breaking down contaminants, and we begin to understand how these communities may be threatened by a warming climate.
Over the past decade, the scientists working on this project have collected numerous microbial communities from the Arctic, including those from seawater, ocean sediments, sea ice, and microbes that live in and on other animals like worms and crustaceans.
We will use next-generation DNA sequencing technologies to probe the diversity and distribution of microbes in these environments, which number many thousands of species. As we learn more about the species diversity, we will then zoom in to focus on the innumerable functional roles played by microbes using metagenomic sequencing. This will allow us to chart ‘metagenomic maps’ that lay the groundwork for a great variety of future studies on the evolutionary paths, ecological adaptations, and ecosystem functions that make Arctic microorganisms fascinating, unique, and fragile.
The primary goals of this project fall into three phases:

Scientists gather samples from a sea ice melt pond in the middle of the Arctic ‘night,’ during a 2005 expedition. During this project, scientists will analyze microbes from samples such as these. Image courtesy of The Hidden Ocean, Arctic 2005 Exploration, NOAA-OE. Download larger version (jpg, 3 MB).
The project is currently in the first phase, data generation. The PIs have amassed several hundred samples collected from around the Arctic, including seawater, sea ice, sediment, and host-associated communities.
These samples are currently undergoing DNA extraction, the first step towards data generation, by graduate student and several undergraduate assistants.
After DNA has been extracted and quality controlled from each sample, a PCR (polymerase chain reaction) amplification step is used to raise the abundance of hypervariable ‘signature’ regions within the small subunit ribosomal RNA genes of Bacteria, Archaea, and Eukaryotes.
These amplified samples will be tagged with unique barcodes, combined, and sequenced on an in-house Illumina MiSeq DNA sequencing machine to a depth of approximately 50,000 sequences per sample. This depth of study will provide an unprecedented look into the diversity of microbial communities in these environments, many of which have never been subjected to next-generation sequencing technologies.
Following the first project phase, collected data will be analyzed to investigate geographic trends, ecological interactions, and overall diversity of the microbial communities in the Arctic.
A small subset of these samples will be selected for additional metagenomic and metatranscriptomic sequencing to maximize the representation of novel microbial species and environments in our datasets.
Unamplified DNA (for metagenomics) and reverse-transcribed RNA (for metatranscriptomics) will be sent to an off-site facility for sequencing on an Illumina HiSeq machine to a depth of about 50,000,000 sequences per sample. This will allow us to investigate the genetic potential (DNA) encoded by the microbial communities, in addition to the subset of that potential that is actively expressed at any given time (RNA).
A number of analyses are planned to investigate genes encoding cold and salt adaptation, oil-degradation potential, and fatty-acid synthesis. The data will be visualized as ‘metagenomics maps’ using self-organizing map algorithms.
The final stage of the project involves synthesis of the huge amounts of data (over 200 billion basepairs) into a number of manuscripts describing the diversity, distribution, ecology, and evolution of the hidden microbial communities found in Arctic marine environments.