
By Eric Collins, Co-Expedition Principal Investigators, University of Alaska Fairbanks
About 50 years ago, two scientists – Linus Pauling and Emile Zuckerkandl – pioneered a new way to categorize life, which has since revolutionized our understanding of how life on Earth is connected.
What Zuckerkandl and Pauling realized was that all life contains information-carrying molecules that can be used to reconstruct the relatedness of organisms. Instead of relying on visible characteristics, like number of teeth or the presence of hair, scientists could directly observe the relatedness between two organisms by comparing their DNA or protein sequences.
DNA is a double helix formed by base pairs attached to a sugar-phosphate backbone. Image courtesy of U.S. National Library of Medicine. Download larger version (jpg, 81 KB).
DNA is the hereditary material that encodes all of the basic functions of life. The basic unit of genetic material is the ‘chromosome,’ which is composed of a long string of many ‘genes.’ When these genes are ‘expressed,’ they are translated into proteins by a complex cellular machine called a ‘ribosome,’ which binds amino acids together in a unique order to make a ‘protein.’ The sequence of a protein is determined by the sequence of the DNA in the gene that encodes it.
Proteins are made of long chains of amino acids that fold into useful forms – the final shape (and function) of the protein depends on its sequence. Because each protein has a unique sequence of amino acids, the same type of protein from two closely related organisms will have similar sequences of amino acids because they derive from a relatively recent common ancestor. The more distantly two organisms are related, the less similarity there will be between their DNA or amino acid sequences. This allows us to establish a system of relatedness between any two organisms that is dependent on the time elapsed since their divergence – this is called ‘molecular phylogenetics.’
Molecular phylogenies are extremely useful because they allow us to categorize life on Earth in a natural way. Instead of relying on the limited number of characteristics that we can visibly observe, we can instead infer relatedness directly. This idea becomes especially important when we are interested in the relatedness of organisms that have very few visible distinguishing characteristics – like microbes.
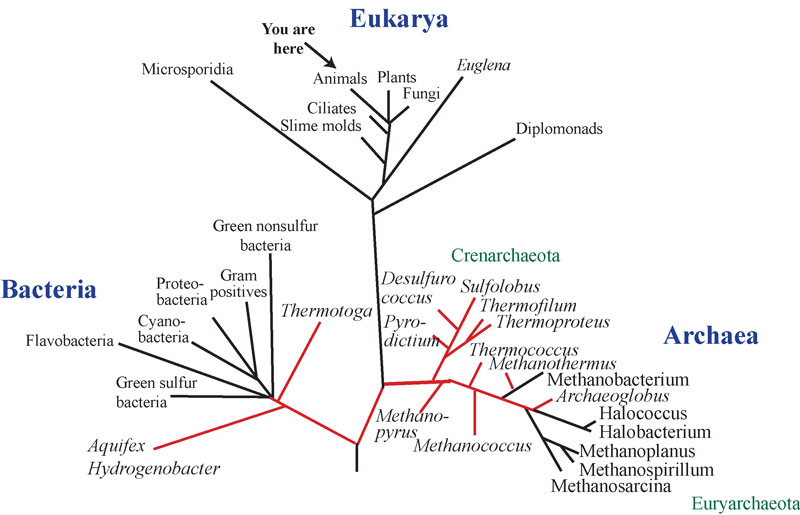
All of life on Earth is divided into three major domains: the Bacteria, Archaea and Eukarya. This is the universal phylogenetic tree based on a gene that we all have, the ribosomal RNA. We (humans) are located in the crown group of animals, but the majority of diversity on this planet is in the microbial (bacterial and archaeal) world. Image adapted from Woese et al. 1990. Download larger version (jpg, 1 MB).
In the 1970s, a biologist named Carl Woese meticulously catalogued the diversity of microbial life using DNA sequences. What he and his colleagues found was that the kingdom Monera, though consisting entirely of single-celled organisms, was in fact composed of two fundamentally different forms of life – now known as Bacteria and Archaea. This finding revolutionized our view of the biological world because it became clear that these were not just new kingdoms to go alongside the existing ones, but made up an even higher order organization of life, which is now known as the ‘Domain.’ Simultaneously, the existing kingdoms of Fungi, Animalia, Plantae, and Protista were shown to be subsets of a domain called Eukarya. This highest ordering of life is now known as the ‘Three Domains’ classification of life.
A remarkable outcome of this re-organization is the finding that the two single-celled domains, Bacteria and Archaea, are not each other’s closest relatives. In fact, it appears most likely that Eukarya (to which humans belong) and Archaea share a more recent common ancestor than do Archaea and Bacteria. These divisions, which are extremely ancient (about three billion years ago), teach us about the early history of life on Earth.
Additional research has further shown that the kingdom Protista is not a ‘monophyletic’ group, that is, the members do not uniquely share a common ancestor. Instead, there are many, perhaps dozens, of kingdoms of single-celled eukaryotes that have arisen independently over time. This further points to the huge unknown biodiversity among microorganisms on Earth and the importance of exploring the hidden realm of microbes.
Recent technological advances in DNA sequencing capabilities have enabled another revolution in environmental microbiology. While it took Woese years to obtain a handful of DNA sequences for his microbes, a modern DNA sequencing machine can reveal an entire human genome sequence in a matter of hours. This is approximately a one-billion fold increase in efficiency over the course of 40 years. The costs have decreased comparably, and it is now possible to obtain billions of basepairs of DNA sequencing data for only a few hundred dollars.
In this project, we’re making use of next-generation DNA sequencing technologies to investigate the diversity of Bacteria and Archaea in the Arctic – in many cases for the first time.
Please follow along for future updates in which we will discuss DNA sequencing technologies in more detail, and explain how, by delving deeply into the diversity of these microorganisms, we hope to learn more about the roles they play in maintaining ecological balance in this fragile and beautiful ecosystem.